miR-375
Introduction
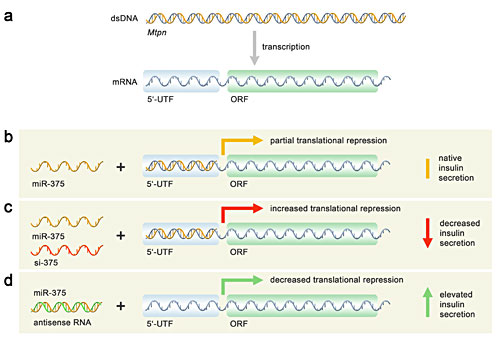
miR-375, an islet specific microRNA, was first discovered by by a team of researchers from Rockefeller University, Lund University (Sweden), New York University, and Oxford University and published at Nature in Nov. 2004. The mechanism by which secretion
is modified by miR-375 is independent of changes in glucose metabolism or intracellular Ca2+-signalling but correlated with a direct effect on insulin exocytosis. Myotrophin (Mtpn) was predicted to be and validated as a target of miR-375. Inhibition of Mtpn by small interfering (si)RNA mimicked the effects of miR-375 on glucosestimulated insulin secretion and exocytosis. Thus, miR-375
is a regulator of insulin secretion and may thereby constitute a novel pharmacological target for the treatment of diabetes [1].
|
|
Panel 1. Translational repression of myotrophin by miR-375. - (a) The dsDNA gene for myotrophin (Mtpn) and its transcribed mRNA, showing both the 5?unstranlated region (UTF) and open reading frame (ORF). (b) In the wild type setting, the single-stranded miR-375 microRNA anneals to a portion of the 5?UTF and represses downstream translation. (c) Upon the addition of small interfering (si) RNAs (siRNA), specifically ones (si-375) that are sequence analogues to miR-375, the effective concentration of miR-375 increases and anneals to more Mtpn 5?UTFs, further repressing Mtpn expression. However, when an antisense RNA (antisense to miR-375) is added, it anneals to miR-375 and prevents it from annealing to the 5?UTR, thereby enhancing Mtpn expression. Mtpn is a component of insulin exocytosis. |
Query Summary
| Query | Type | Family Accession | Family ID | miRBase ID1 | miRBase ID2 | # of members |
| mir-375 | family | MIPF0000114 | mir-375 | . | . | 8 |
microRNA family : mir-375 [MIPF0000114] miRBase
| ID | Accession | Chromosome | Start | End | Strand | Genome | MIRANDA1 | TARGETSCAN1 | MIRANDA2 | TARGETSCAN2 | Structure |
| dre-mir-375-1 | MI0002072 | 6 |
13423599 |
13423678 |
- |
Coordinate | dre-miR-375 | dre-mir-375-1 | |||
| dre-mir-375-2 | MI0002073 | 9 |
10232524 |
10232606 |
- |
Coordinate | dre-miR-375 | dre-mir-375-2 | |||
| fru-mir-375 | MI0003419 | scaffold_3113 scaffold_83 |
9349 92347 |
9425 92423 |
+ + |
Coordinate | fru-miR-375 | fru-mir-375 | |||
| gga-mir-375 | MI0003705 | 7 |
23117692 |
23117756 |
+ |
Coordinate | gga-miR-375 | gga-miR-375 | gga-mir-375 | ||
| hsa-mir-375 | MI0000783 | 2 |
219574611 |
219574674 |
- |
Coordinate | hsa-miR-375 | hsa-miR-375 | hsa-mir-375 | ||
| mdo-mir-375 | MI0005327 | 7 |
175492651 |
175492713 |
+ |
Coordinate | mdo-miR-375 | mdo-mir-375 | |||
| mmu-mir-375 | MI0000792 | 1 |
74833865 |
74833928 |
- |
Coordinate | mmu-miR-375 | mmu-miR-375 | mmu-mir-375 | ||
| tni-mir-375 | MI0003420 | 2 |
15358681 |
15358757 |
- |
Coordinate | tni-miR-375 | tni-mir-375 |
microRNA stem-loop sequence alignment
CLUSTAL W (1.83) multiple sequence alignment
hsa-mir-375 --------CCCCGCGACGAGCCCCUCGCACAA-ACCGG-ACCUGAGC--GUUUUGUUCGU
mmu-mir-375 --------CCCCGCGACGAGCCCCUCGCACAA-ACCGG-ACCUGAGC--GUUUUGUUCGU
mdo-mir-375 ---------CCCGCGCCGAGCCCCUCGCACAA-ACCGG-ACCUGAGC--GUUUUGUUCGU
gga-mir-375 --------CCUGGCGUCGAGCCCCACGUGCAAGACCUG-ACCUGAAC--GUUUUGUUCGU
fru-mir-375 UGUACUUGCUUCACGUUGAGCCUCACGCACAAUACCUGAAGAUGAA---GUUUUGUUCGU
tni-mir-375 UGUACUUGCUUCACGUUGAGCCUCACGCACAAUACCUGAAGAUGAA---GUUUUGUUCGU
dre-mir-375-1 UGCACUUGCUUUACGUUGAGCCACACGCACAAUACAUGUGGAUUCA---GUUUUGUUCGU
dre-mir-375-2 UGUACUUGUCUCACGUUGAGCCACACGCACAAUGCCUGCAGAUGAAAGGGUUUUGUUCGU
** ***** * ** *** * * * ***********
hsa-mir-375 UCGGCUCGCGUGAGGC-------
mmu-mir-375 UCGGCUCGCGUGAGGC-------
mdo-mir-375 UCGGCUCGCGUGAGGC-------
gga-mir-375 UCGGCUCGCGUUAGGC-------
fru-mir-375 UCGGCUCGCGUUAUGCAGGU---
tni-mir-375 UCGGCUCGCGUUAUGCAGGU---
dre-mir-375-1 UCGGCUCGCGUUAAGCAAGUGCA
dre-mir-375-2 UCGGCUCGCGUUACGCAGAUGCA
*********** * **

Conserved Secondary Structure Prediction - By RNAalifold in ViennaRNA package
________CCUCACGUCGAGCCCCACGCACAAUACCUG_ACAUGAAC__GUUUUGUUCGUUCGGCUCGCGUUAGGC_______
............(((.((((((...((.((((.((..............)).)))).))...)))))))))............ (-17.55 = -16.83 + -0.72)
............(((.((((((...((.((((.((..............)).)))).))...)))))))))............ [-17.96]
frequency of mfe structure in ensemble 0.507898